TRACK-TBI NET ICU Working Group
Last month, we met with the TRACK-TBI NET ICU Working Group Monthly Meeting to discuss ongoing projects and research opportunities with multimodal neuromonitoring data. We showcased the Moberg Sandbox environment that allows users of the Moberg Cloud Platform to access multimodal data using JupyterHub with a variety of programming tools, such as Python, Julia, and R (see this link for a full list of available kernels). Several of the working group members have already requested access to the platform to explore the data and start their analyses.
If you are a part of the TRACK-TBI NET ICU Working Group (or you have interest in joining), please contact Dr. Brandon Foreman (foremabo@ucmail.uc.edu) from the University of Cincinnati for access to this environment.
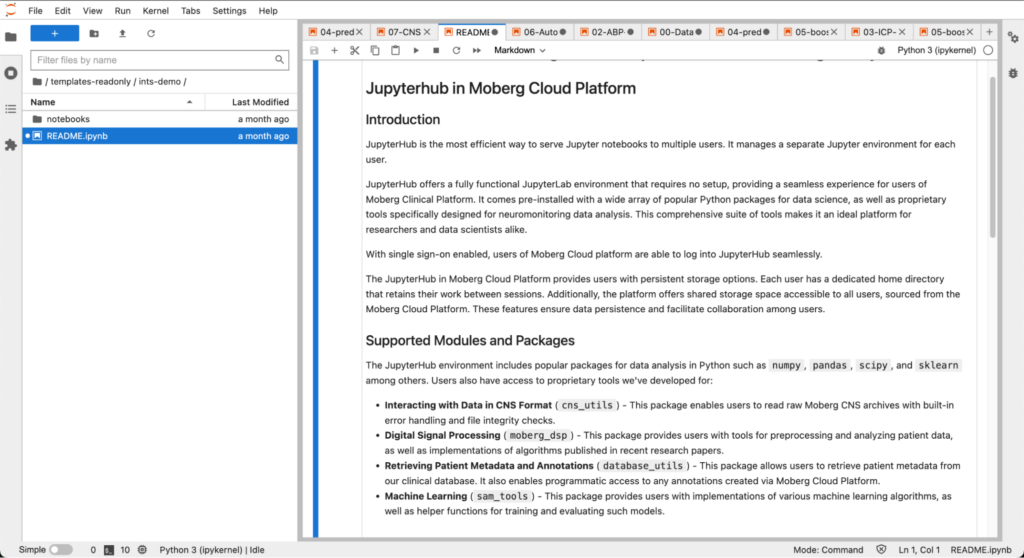
The Moberg Sandbox has many data science and signal processing libraries pre-installed. It also comes equipped with libraries that we have developed for reading data from the CNS Monitor, processing multimodal signals, retrieving patient metadata from a relational database, and training and testing machine learning algorithms.
Moberg Sandbox Overview
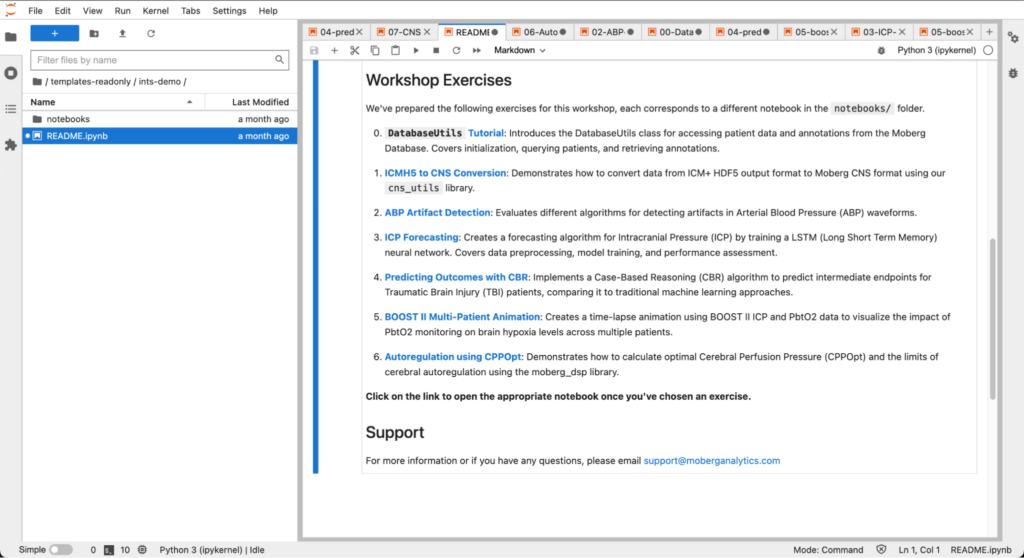
We have included several exercises as example:
- How to access patient metadata from our relational database
- How to convert data from ICM+ exported as Hierarchical Data Format 5 (HDF5) to the CNS format to interoperate between ICM+ and our platform
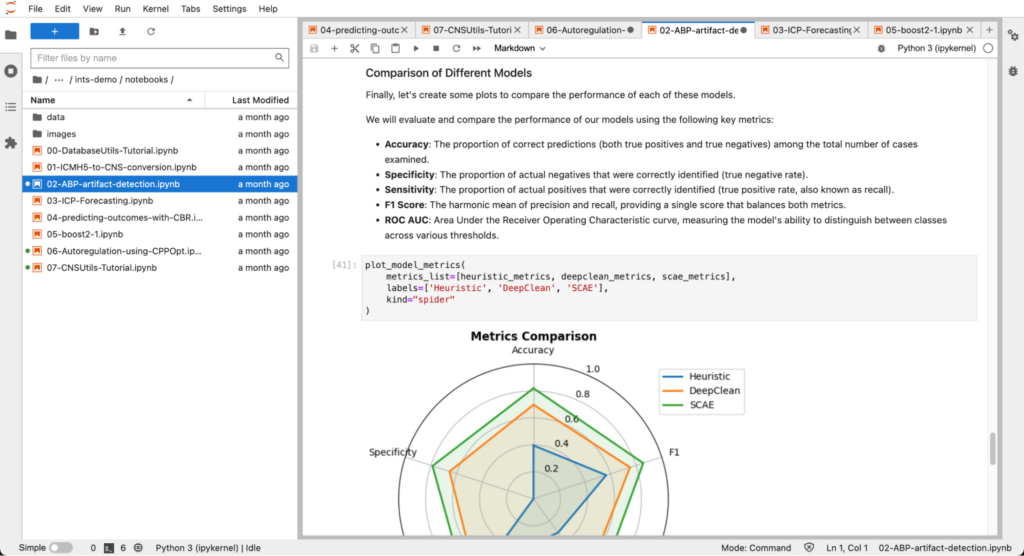
- How to design, train, and test models to detect artifacts in the arterial blood pressure (ABP) waveform collected from the ICU
- How to design, train, and test models to perform intracranial pressure (ICP) time series forecasting
- How to design, train, and test models for prognostication of short and long term outcomes
- How to build a multi-patient animation showing the evolution of patient status over time
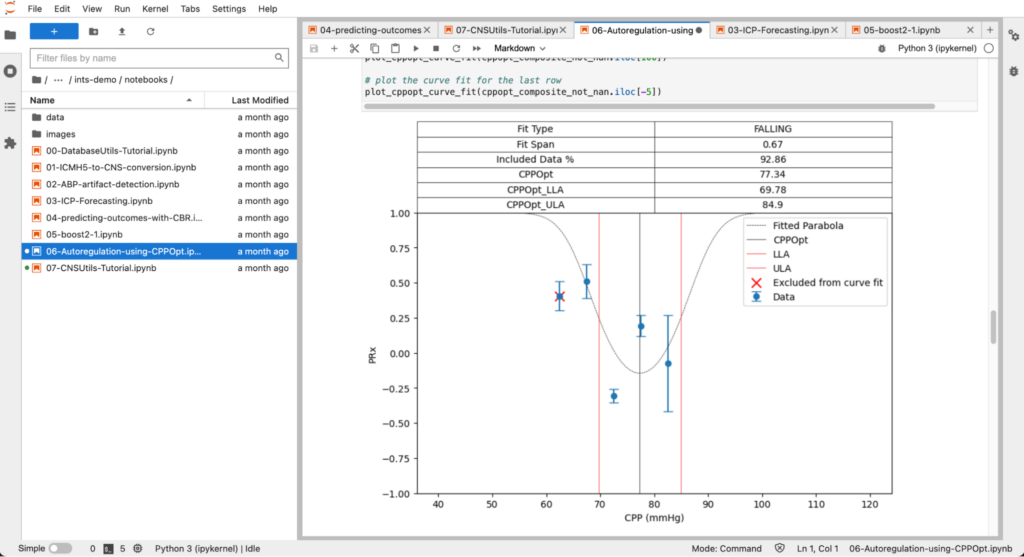
- How to calculate PRx and CPPopt from ICP and ABP